NCC presenting the success story
NCC Spain
💡A new Success Story leveraging #HPC to design anti-cancer drugs💡
📋 "Simulation and modeling of biosystems with applications to medicine: exploring the structural diversity of the MYC protein and its implications for in-silico design of anti-cancer drugs" led by Jordi Marti Rabassa from Universitat Politècnica de Catalunya (UPC)
The MYC protein is a master regulator of cell growth, proliferation, and metabolism. Its overexpression or dysregulation is indicative of numerous cancers, including breast, colon, lung, and haematologic cancer. Despite its well-established oncogenic role, MYC has historically been labelled “difficult-to-drug” due to its structure and mechanism of action.
This is largely due to its intrinsically disordered nature, a structural feature that sets it apart from classical drug targets and drug design approaches. For this reason, simulation and modelling becomes a promising frontier for developing novel, structure-guided anti-cancer therapies.
🖥️ Thanks to RES supercomputer #MareNostrum5Acc from Barcelona Supercomputing Center, the team performed 3000ns of Molecular Dynamics (MD) and 5000ns of Well-Tempered Metadynamics (WTM) simulations of the MYC in solution.
CLIENT/USER PROFILE:
Researchers and scientists in the field of cancer biology, structural biology, and computational chemistry, particularly those involved in the study of the MYC protein and the development of anti-cancer therapies.
IMPACT:
The project has the potential to significantly impact the development of new anti-cancer therapies by providing a well-defined and stable 3D structure of the MYC protein, which is a challenging target due to its intrinsically disordered nature.
BENEFITS:
Advancements in MYC protein research: The project provides valuable insights into the structural diversity of the MYC protein, which is a critical step towards understanding its function and developing effective anti-cancer therapies.
Structure-based drug design: The results of the project enable the use of structure-based drug design strategies to identify potential binding pockets and design novel anti-cancer therapies.
Advancements in computational biology: The project demonstrates the power of high-performance computing (HPC) in tackling complex biological systems and provides a valuable resource for the computational and medicinal chemistry communities.
KEY POINTS BEFORE AGREEING ON THE PROJECT:
Clear objectives: Defining the project's goals, including the specific research questions to be addressed and the expected outcomes.
Computational resources: Ensuring access to sufficient computational resources, such as the MareNostrum5 supercomputer, to perform the required simulations.
Interdisciplinary collaboration: Collaborating with experts from various fields, including cancer biology, structural biology, and computational chemistry, to ensure the project's success.
TECHNICAL/SCIENTIFIC CHALLENGE:
The project faced the challenge of understanding the complex and dynamic structure of the MYC protein, which is intrinsically disordered and challenging to study using traditional structural biology techniques.
SOLUTION:
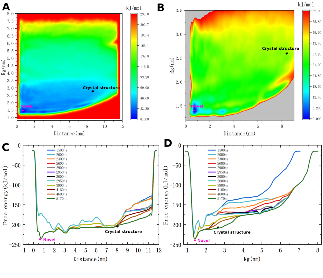
The team utilized Molecular Dynamics and Well-Tempered Metadynamics simulations on the MareNostrum5 supercomputer to capture the most stable folded conformation of the MYC protein in its unbound state. The results provided a well-defined and stable 3D structure of MYC, which is a critical step towards the development of novel anti-cancer therapies.
They captured the most stable folded conformation of the MYC protein in its unbound state, which is highly relevant given its intrinsical disorder and its "undruggable" status. Currently, the team is performing other MYC-MAX and MYC-MAX-DNA simulations.
The results of the simulations provided, for the first time, a well-defined and stable 3D structure of MYC in its free form. This breakthrough offers new opportunities to identify persistent, druggable binding pockets and opens the door to structure-based drug design strategies against one of the most important but hard-to-target proteins in cancer, representing:
🔹A valuable resource for computational and medicinal chemistry communities
🔹A foundational step towards rational design of anti-cancer therapies
🔹A demonstration of the power of #HPC in tackling biologically complex and pharmacologically resistant systems.
📸 Results from WTM simulations show original and zoomed-in free energy landscapes (A-B) and free-energy profiles along one collective variable (C-D). Crystal structures and the captured stable folded structures are indicated using stars in black and purple.