TITLE: Structure and recognition mechanism of the HIV-1 Rev response element revealed by mutagenesis and FRET-monitored modeling.
https://www.sciencedirect.com/science/article/pii/S0022283625005170?via%3Dihub
IMPACT
Understanding the atomic-level structure of the HIV-1 RRE (Rev Response Element) and its recognition by the Rev protein has profound implications for antiviral research. By revealing how this RNA–protein complex assembles—a process essential for exporting unspliced viral RNA from the nucleus—this work directly targets a critical step in HIV-1 replication. The resulting structural insights create new opportunities for therapeutic intervention in a system that has remained largely inaccessible due to the absence of high-resolution data.
BENEFITS
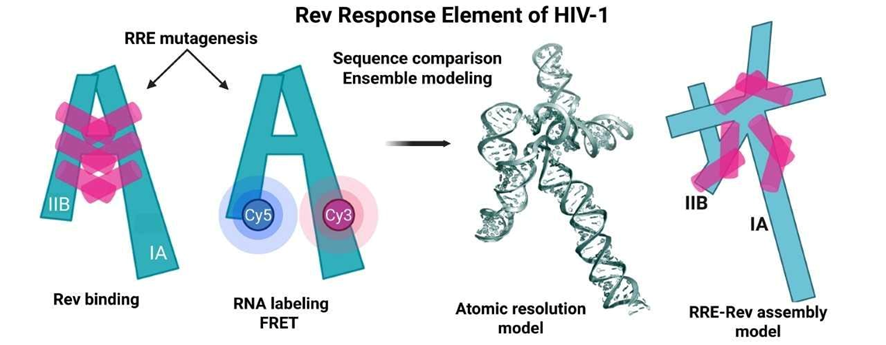
The study provides an atomic model of the RRE that clarifies both its three-dimensional architecture and the mechanism by which it assembles with Rev.
This knowledge:
Opens new directions for designing drugs that disrupt the RRE–Rev complex, thereby inhibiting a key stage in HIV-1 replication.
Identifies specific structural features of the RRE as potential therapeutic targets.
Offers a foundation that could improve or complement current antiretroviral treatments by focusing on RNA–protein interactions previously difficult to target.
KEY POINTS BEFORE AGREEING ON THE PROJECT
Before committing to the project, it is essential to consider:
The need for advanced experimental tools capable of probing RNA structure and dynamics.
The availability of expertise in fluorescence-based measurements, mutagenesis, and computational modeling.
The complexity of interpreting RNA conformational changes, which can require iterative cycles of experimentation and modeling.
The importance of validating proposed atomic models through multiple complementary techniques.
The significant time and computational resources required for all-atom structural modeling.
TECHNICAL/SCIENTIFIC CHALLENGE
The central challenge is that the three-dimensional structure of the HIV-1 RRE and its precise mode of recognition by the Rev protein remain unknown at atomic resolution. This lack of structural information has hindered efforts to develop antiretroviral therapies targeting this system, which is essential for the nuclear export of unspliced viral RNA. The project therefore requires overcoming the intrinsic complexity of RNA folding, junction dynamics, and RNA–protein assembly.
SOLUTION
The researchers developed a multi-disciplinary experimental and computational strategy that integrates:
FRET (Förster Resonance Energy Transfer) to measure distances between domains of the RRE.
Targeted mutagenesis to alter key regions of the RNA and evaluate their structural contributions.
All-atom computational modeling to generate three-dimensional structures consistent with the experimental data.
This combined approach allowed the identification of how three-way and four-way junctions within the RRE determine its spatial organization and influence the formation of the RRE–Rev complex.